It began about 12 years ago when research Professor Paul Hebert proposed using short sections of DNA to identify species.
To simplify understanding, it’s being called barcoding of species, in that species can be identified through computer matching of the DNA codes, similar to the way product barcodes are read by store scanners.
The idea has been gaining momentum, and was among the recommendations this week to help identify and aid in preservation of species at the UN’s Biodiversity Conference in Cancun, Mexico.
Paul Hebert (O.C., PhD, FRSC) is the Director for the Centre of Biodiversity Genomics at the University of Guelph in Ontario.
Listen
Professor Hebert proposed the idea in 2002 in a scientific paper to the Royal Society called “Biological Identification through DNA barcodes”
The idea was accepted among the recommendations this week as representatives from 196 countries attended the 13th conference of the Parties to the Convention on Biological Diversity (CBD).
This Canadian originated idea is now part of the United Nations strategic plan for enhancing and protecting biodiversity.
Malcolm Campbell, vice-president (research) at the university said, “Because of the visionary work of U of G researchers, DNA barcoding has changed the way we look at life on this planet. The UN’s recognition is a testament to its effectiveness and the important role it plays in protecting the world’s biodiversity.”
As professor Hebert points out, it would be impossible to track temperature and climate without instruments to measure, and this is an instrument to track species to determine, their extent and changes in biodiversity, their growth or decline and effects of environmental change.

He also points out that determining the species in a lake can now simply be done not by catching specimens, but by simply taking a water sample and identifying the traces of DNA in it, from fish, to algae and so on.
In addition only a specific region of the DNA is needed, a short section known as C01. Because it is short, it is relatively quick and inexpensive to sequence yet provides enough information to clearly identify species.
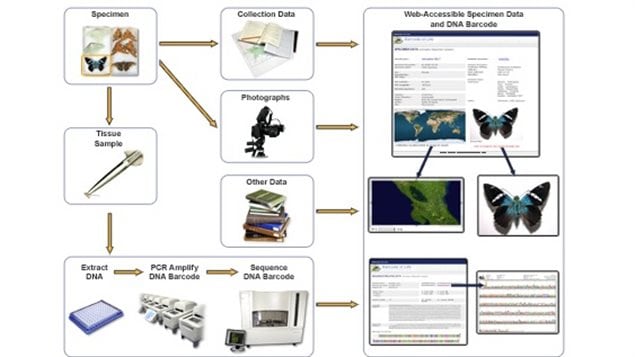
Once a sample tissue or DNA is collected it is analyzed and sequenced. In written form the code is represented by a series of letters CATG representing the nucleic acids – cytosine, adenine, thymine and guanine.

The registry project now involves scientists from around the world who constantly add and request information from the Guelph-based registry.
Additional information- sources







For reasons beyond our control, and for an undetermined period of time, our comment section is now closed. However, our social networks remain open to your contributions.